2.2.4
Linear Discriminant Analysis (LDA)
Summary
- LDA finds directions that maximize the ratio of between-class variance to within-class variance, serving both classification and dimensionality reduction.
- The decision boundary takes the form \(\mathbf{w}^\top \mathbf{x} + b = 0\), which becomes a line in 2D or a plane in 3D, giving a clear geometric interpretation.
- Assuming each class is Gaussian with the same covariance matrix, LDA approaches the Bayes-optimal classifier.
- scikit-learn’s
LinearDiscriminantAnalysismakes it easy to visualise decision boundaries and to inspect the projected features.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
Mathematical formulation #
For the two-class case, the projection direction \(\mathbf{w}\) maximizes
$$ J(\mathbf{w}) = \frac{\mathbf{w}^\top \mathbf{S}_B \mathbf{w}}{\mathbf{w}^\top \mathbf{S}_W \mathbf{w}}, $$where \(\mathbf{S}_B\) is the between-class scatter matrix and \(\mathbf{S}_W\) is the within-class scatter matrix. In the multi-class case we obtain up to \(K-1\) projection directions, which can be used for dimensionality reduction.
Experiments with Python #
Below we apply LDA to a synthetic two-class data set, draw the decision boundary, and plot the projected 1D features. Calling transform returns the projected data directly.
| |
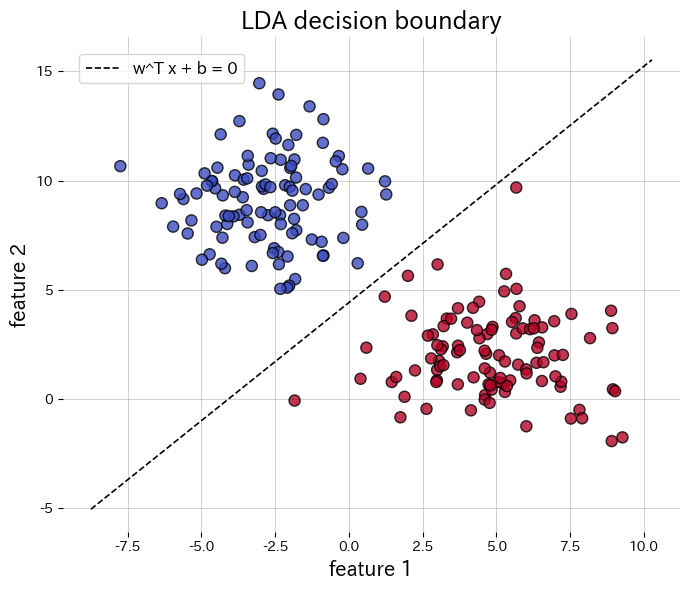
References #
- Fisher, R. A. (1936). The Use of Multiple Measurements in Taxonomic Problems. Annals of Eugenics, 7(2), 179–188.
- Hastie, T., Tibshirani, R., & Friedman, J. (2009). The Elements of Statistical Learning. Springer.