2.5.4
DBSCAN
- DBSCAN (Density-Based Spatial Clustering of Applications with Noise) groups points according to local density so clusters can take any shape while sparse regions become noise.
- Two hyperparameters control the model:
eps, the neighbourhood radius, andmin_samples, the minimum number of neighbours required for a point to become a core sample. - Points are labelled core, border, or noise; clusters are connected components of core points plus their border neighbours.
- A common tuning recipe is to fix
min_samples(≥ dimensionality + 1) and sweepepswhile inspecting the share of points flagged as noise.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
1. Overview #
DBSCAN does not require the number of clusters beforehand. Instead, it inspects each sample:
- Core points: at least
min_samplesneighbours within distanceeps. - Border points: lie within the
eps-ball of a core point but fail the core criterion themselves. - Noise points: belong to no core neighbourhood.
Because of this density view, DBSCAN is robust to non-convex clusters such as two moons or concentric circles. Always scale the features so eps has a meaningful interpretation.
2. Formal definition #
Given (x_i \in \mathcal{X}), its (\varepsilon)-neighbourhood is
$$ \mathcal{N}_\varepsilon(x_i) = \{ x_j \in \mathcal{X} \mid \lVert x_i - x_j \rVert \le \varepsilon \}. $$If (|\mathcal{N}_\varepsilon(x_i)| \ge \texttt{min_samples}|), the point is core. DBSCAN builds clusters by exploring density-reachable points; anything left unvisited becomes noise. With a spatial index the overall complexity is (O(n \log n)).
3. Python example #
The snippet below uses scikit-learn’s DBSCAN on a two-moons dataset, colours core/border points differently, and reports how many samples are marked as noise.
| |
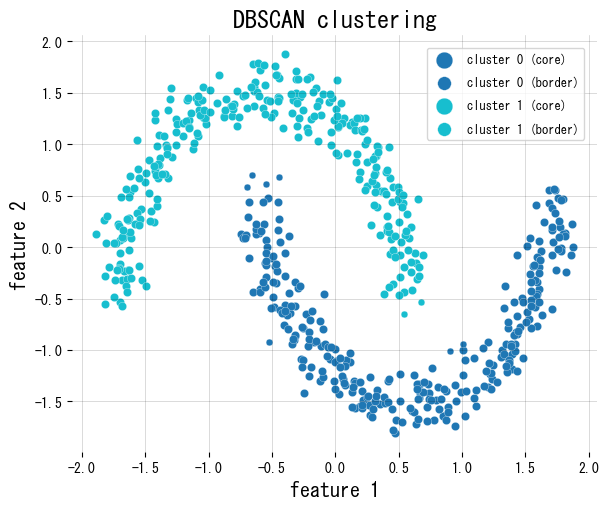
4. Practical tips #
- Plot the sorted distances to the k-th neighbour (k =
min_samples) to pickepswhere the curve exhibits an elbow. - Use pipelines so standardisation and clustering run together; otherwise distance-based decisions become meaningless.
- For very large datasets consider approximate nearest neighbours or switch to HDBSCAN, which extends DBSCAN to multi-density data and removes the need to tune
eps.
5. References #
- Ester, M., Kriegel, H.-P., Sander, J., & Xu, X. (1996). A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. KDD.
- Schubert, E., Sander, J., Ester, M., Kriegel, H.-P., & Xu, X. (2017). DBSCAN Revisited, Revisited. ACM Transactions on Database Systems.
- scikit-learn developers. (2024). Clustering. https://scikit-learn.org/stable/modules/clustering.html