2.5.5
Gaussian Mixture Models (GMM)
Summary
- A Gaussian Mixture Model represents data as a weighted sum of multivariate normal components.
- It outputs a responsibility matrix that quantifies how strongly each component explains every sample.
- Parameters are estimated with the EM algorithm; covariance structures can be
full,tied,diag, orspherical. - Model selection typically combines information criteria (BIC/AIC) with multiple random initialisations for stability.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
Mathematics #
The density of \(\mathbf{x}\) is
$$ p(\mathbf{x}) = \sum_{k=1}^{K} \pi_k \, \mathcal{N}(\mathbf{x} \mid \boldsymbol{\mu}_k, \boldsymbol{\Sigma}_k), $$with mixture weights \(\pi_k\) (non-negative and summing to 1). EM alternates:
- E-step: compute responsibilities \(\gamma_{ik}\). $$ \gamma_{ik} = \frac{\pi_k \, \mathcal{N}(\mathbf{x}_i \mid \boldsymbol{\mu}_k, \boldsymbol{\Sigma}_k)} {\sum_{j=1}^K \pi_j \, \mathcal{N}(\mathbf{x}_i \mid \boldsymbol{\mu}_j, \boldsymbol{\Sigma}_j)}. $$
- M-step: re-estimate \(\pi_k, \boldsymbol{\mu}_k, \boldsymbol{\Sigma}k\) using \(\gamma{ik}\) as weights.
The log-likelihood increases monotonically and converges to a local optimum.
Python walkthrough #
We fit a GMM to synthetic 2D blobs, plot the hard assignments, and report mixture weights and the responsibility matrix shape.
| |
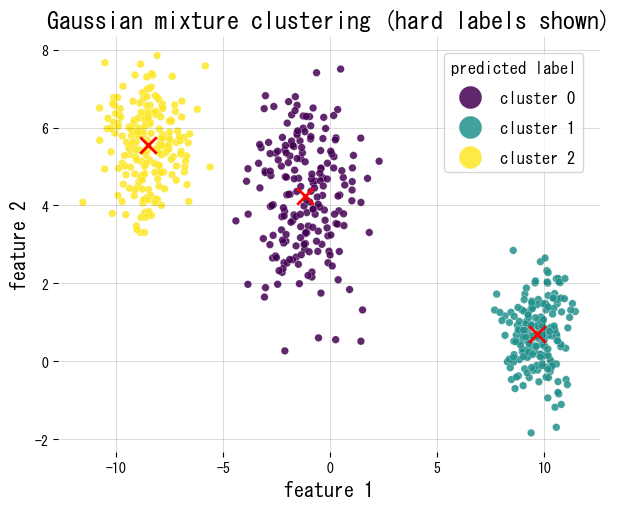
References #
- Bishop, C. M. (2006). Pattern Recognition and Machine Learning. Springer.
- Dempster, A. P., Laird, N. M., & Rubin, D. B. (1977). Maximum Likelihood from Incomplete Data via the EM Algorithm. Journal of the Royal Statistical Society, Series B.
- scikit-learn developers. (2024). Gaussian Mixture Models. https://scikit-learn.org/stable/modules/mixture.html