2.4.1
Random Forests
Summary
- Random Forest combines bootstrap sampling with random feature subsets to build diverse decision trees.
- Aggregating many decorrelated trees reduces variance and usually improves out-of-sample performance.
- Hyperparameters such as
n_estimators,max_features, and tree depth determine the balance between bias, variance, and runtime.
Intuition #
Random Forest is bagging plus feature randomness at each split. Because trees are forced to look at different subsets of predictors, they become less correlated, and averaging them gives a stronger and more robust final model.
Detailed Explanation #
import numpy as np
import matplotlib.pyplot as plt
Train Random Forests #
For ROC-AUC, see ROC-AUC for an explanation of how to plot.
from sklearn.datasets import make_classification
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
n_features = 20
X, y = make_classification(
n_samples=2500,
n_features=n_features,
n_informative=10,
n_classes=2,
n_redundant=0,
n_clusters_per_class=4,
random_state=777,
)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=777
)
model = RandomForestClassifier(
n_estimators=50, max_depth=3, random_state=777, bootstrap=True, oob_score=True
)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
rf_score = roc_auc_score(y_test, y_pred)
print(f"ROC-AUC @ test dataset = {rf_score}")
ROC-AUC @ test dataset = 0.814573097628059
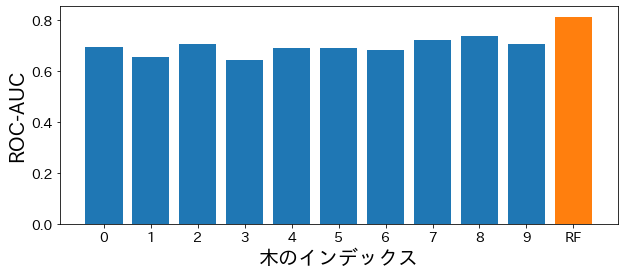
Check the performance of each tree in the random forest #
import japanize_matplotlib
estimator_scores = []
for i in range(10):
estimator = model.estimators_[i]
estimator_pred = estimator.predict(X_test)
estimator_scores.append(roc_auc_score(y_test, estimator_pred))
plt.figure(figsize=(10, 4))
bar_index = [i for i in range(len(estimator_scores))]
plt.bar(bar_index, estimator_scores)
plt.bar([10], rf_score)
plt.xticks(bar_index + [10], bar_index + ["RF"])
plt.xlabel("tree index")
plt.ylabel("ROC-AUC")
plt.show()
Feature Importance #
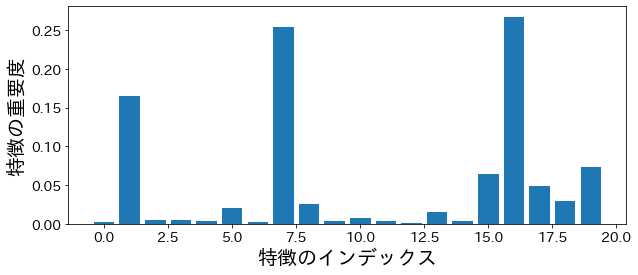
Importance based on impurity #
plt.figure(figsize=(10, 4))
feature_index = [i for i in range(n_features)]
plt.bar(feature_index, model.feature_importances_)
plt.xlabel("Feature Index")
plt.ylabel("Feature Importance")
plt.show()
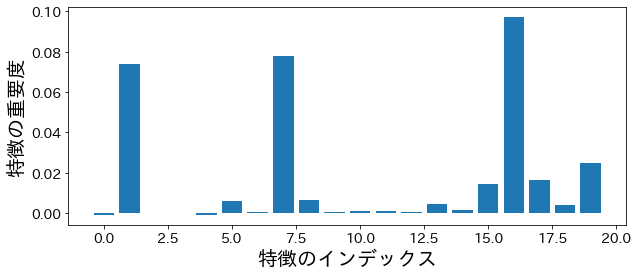
permutation importance #
from sklearn.inspection import permutation_importance
p_imp = permutation_importance(
model, X_train, y_train, n_repeats=10, random_state=77
).importances_mean
plt.figure(figsize=(10, 4))
plt.bar(feature_index, p_imp)
plt.xlabel("Feature Index")
plt.ylabel("Feature Importance")
plt.show()
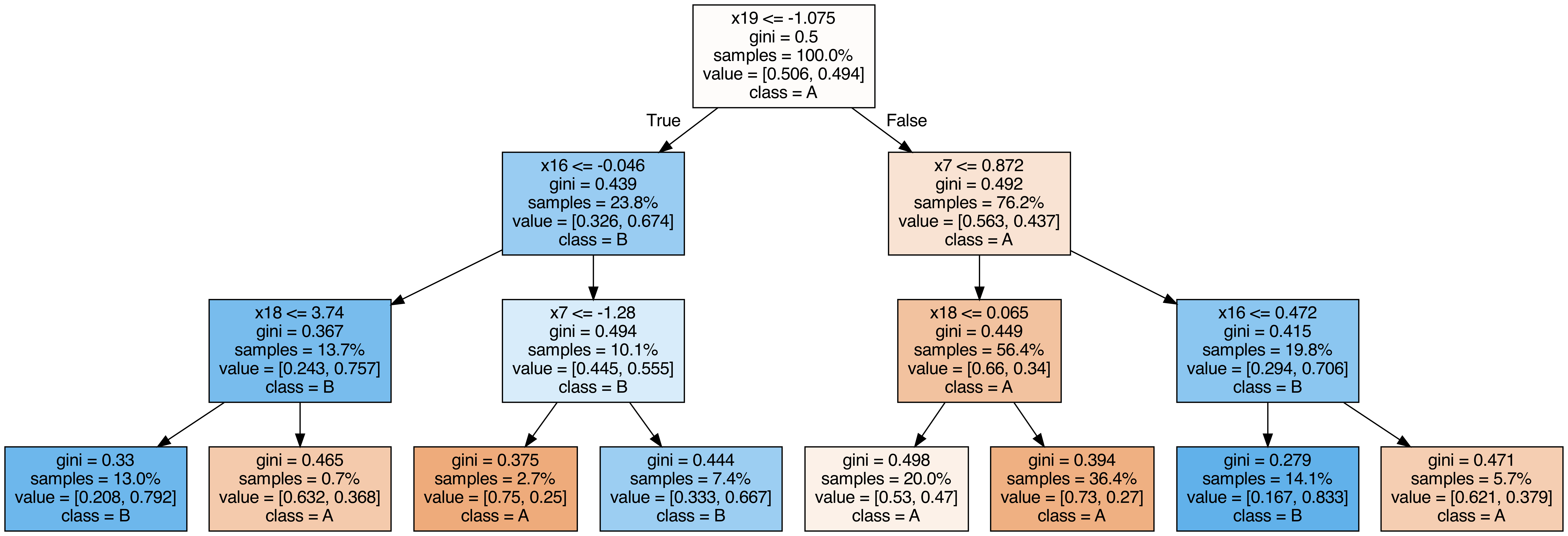
Output each tree contained in the random forest #
from sklearn.tree import export_graphviz
from subprocess import call
from IPython.display import Image
from IPython.display import display
for i in range(10):
try:
estimator = model.estimators_[i]
export_graphviz(
estimator,
out_file=f"tree{i}.dot",
feature_names=[f"x{i}" for i in range(n_features)],
class_names=["A", "B"],
proportion=True,
filled=True,
)
call(["dot", "-Tpng", f"tree{i}.dot", "-o", f"tree{i}.png", "-Gdpi=500"])
display(Image(filename=f"tree{i}.png"))
except KeyboardInterrupt:
# TODO
pass
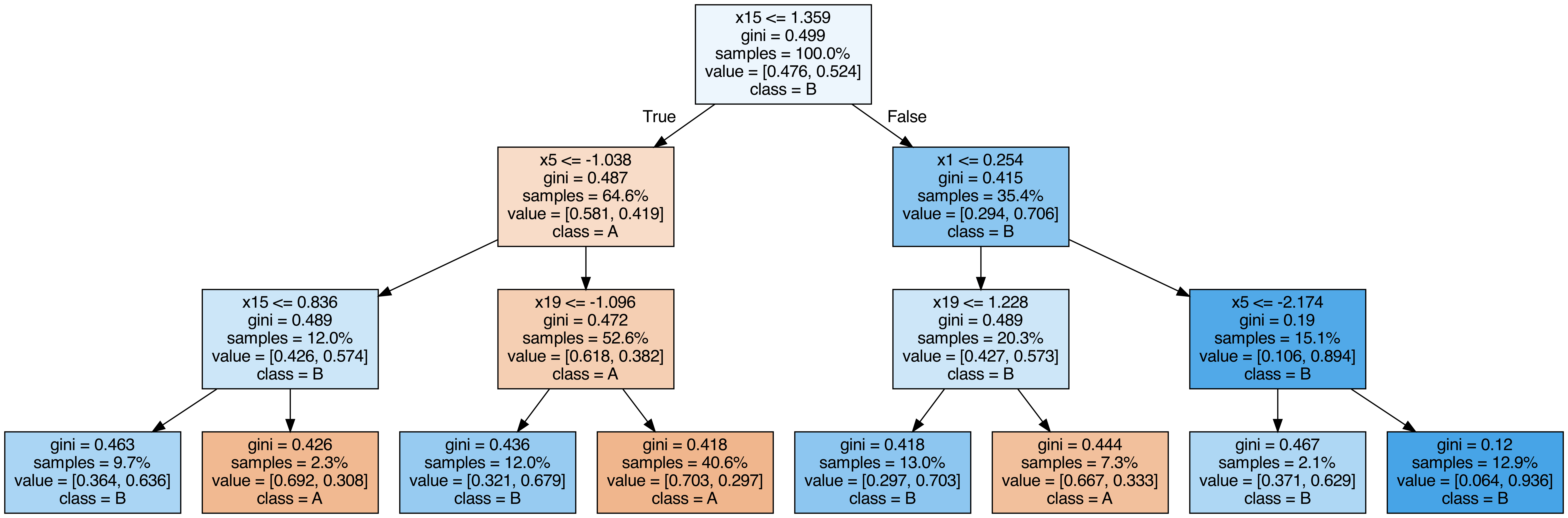
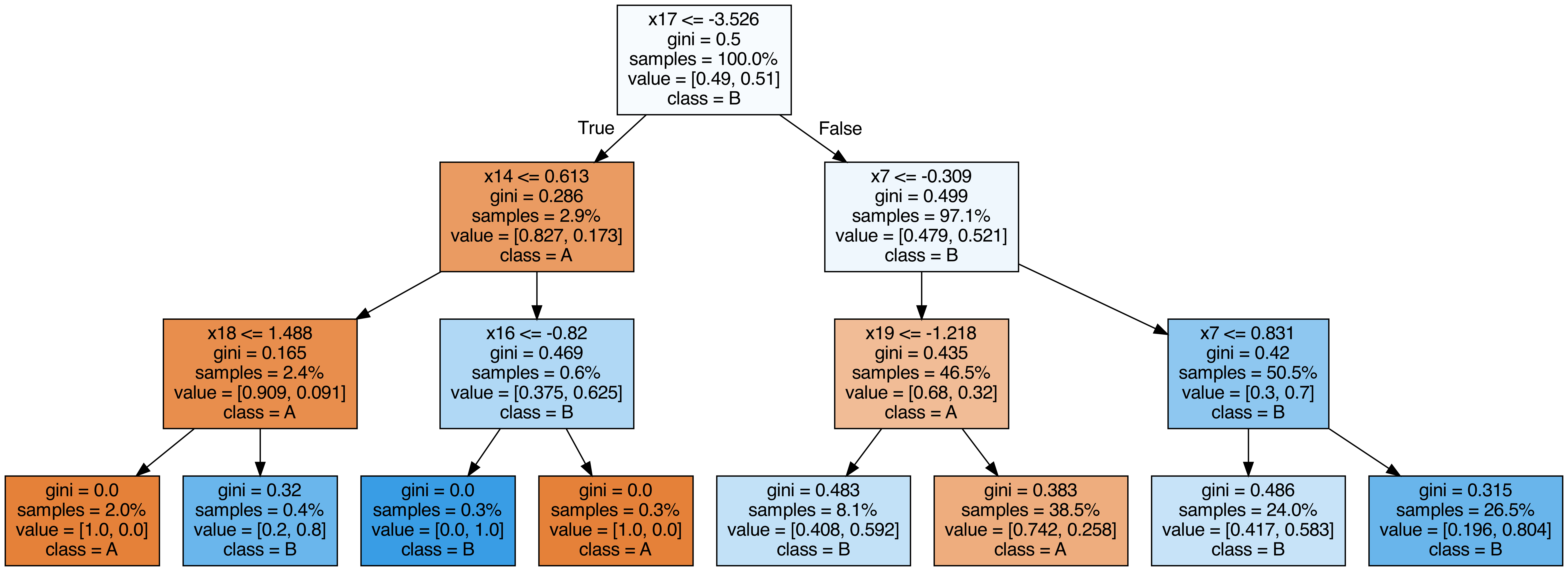
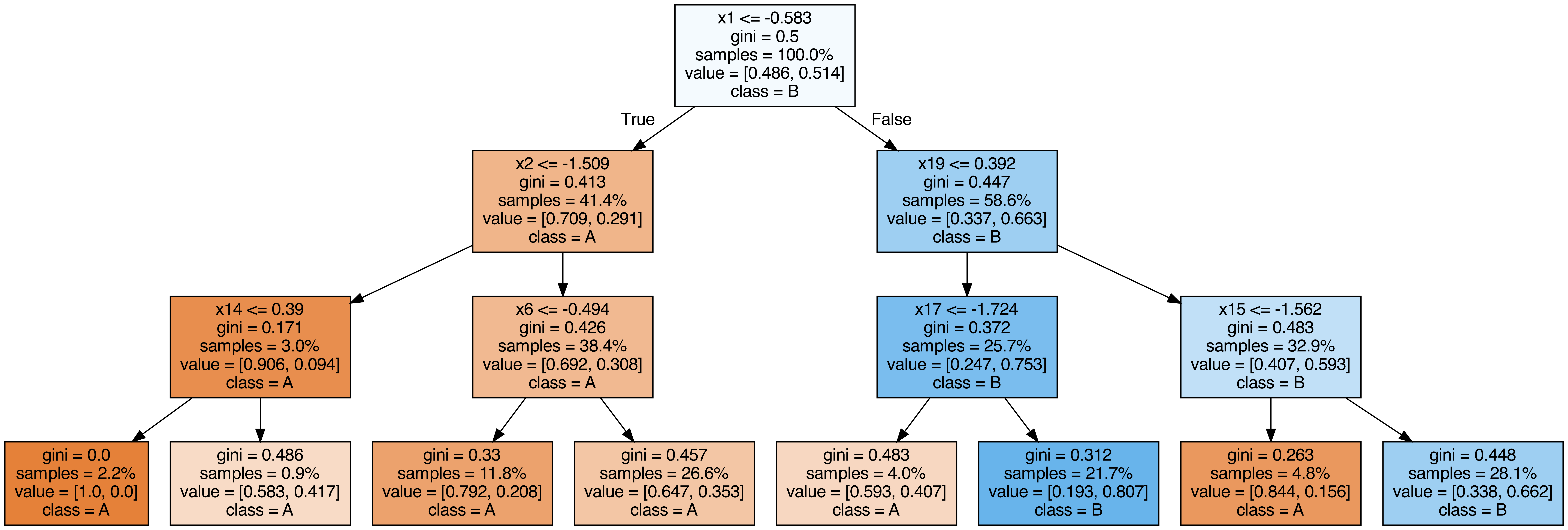
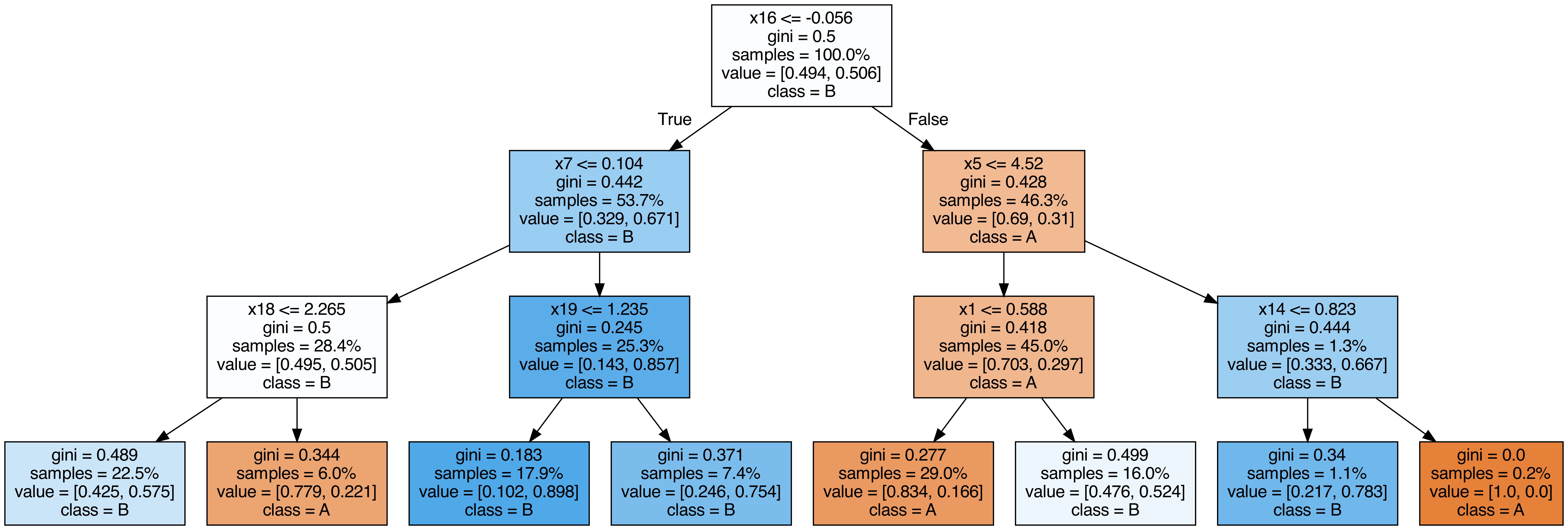
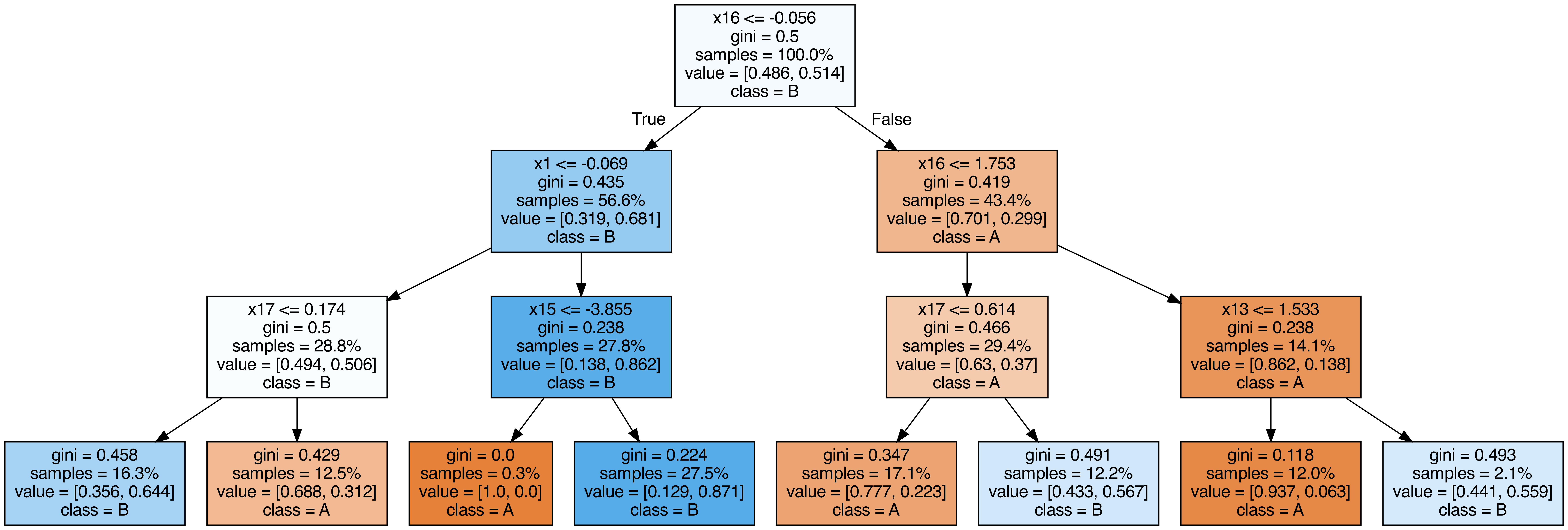
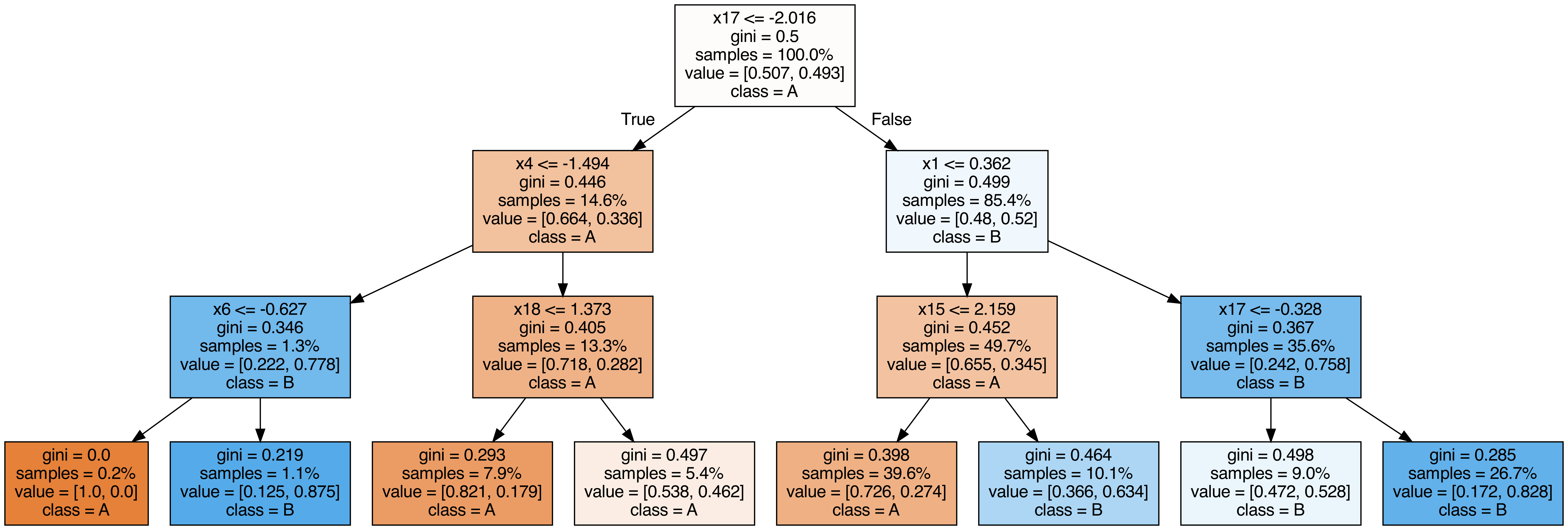
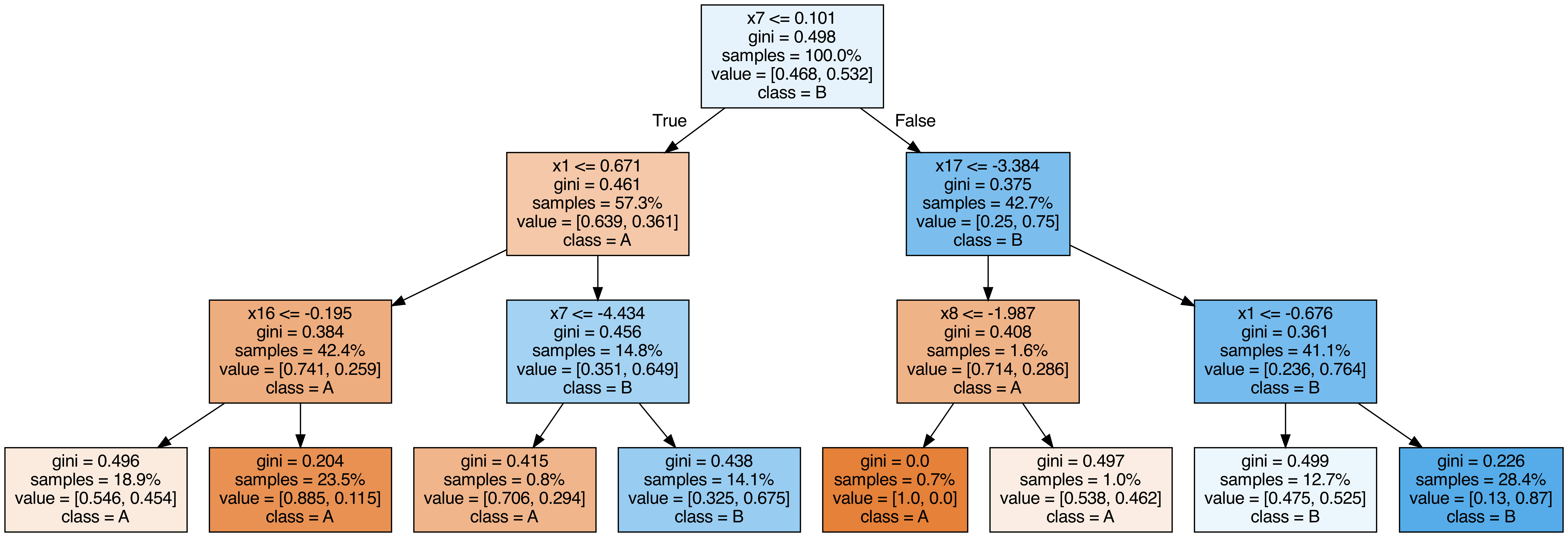
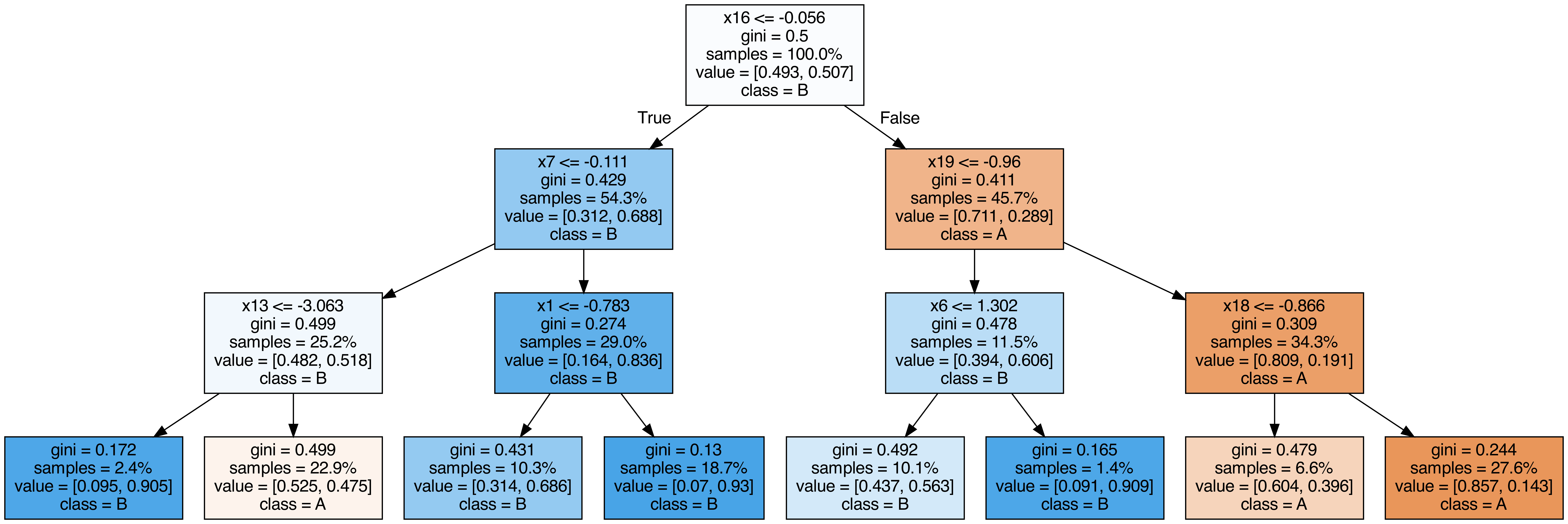
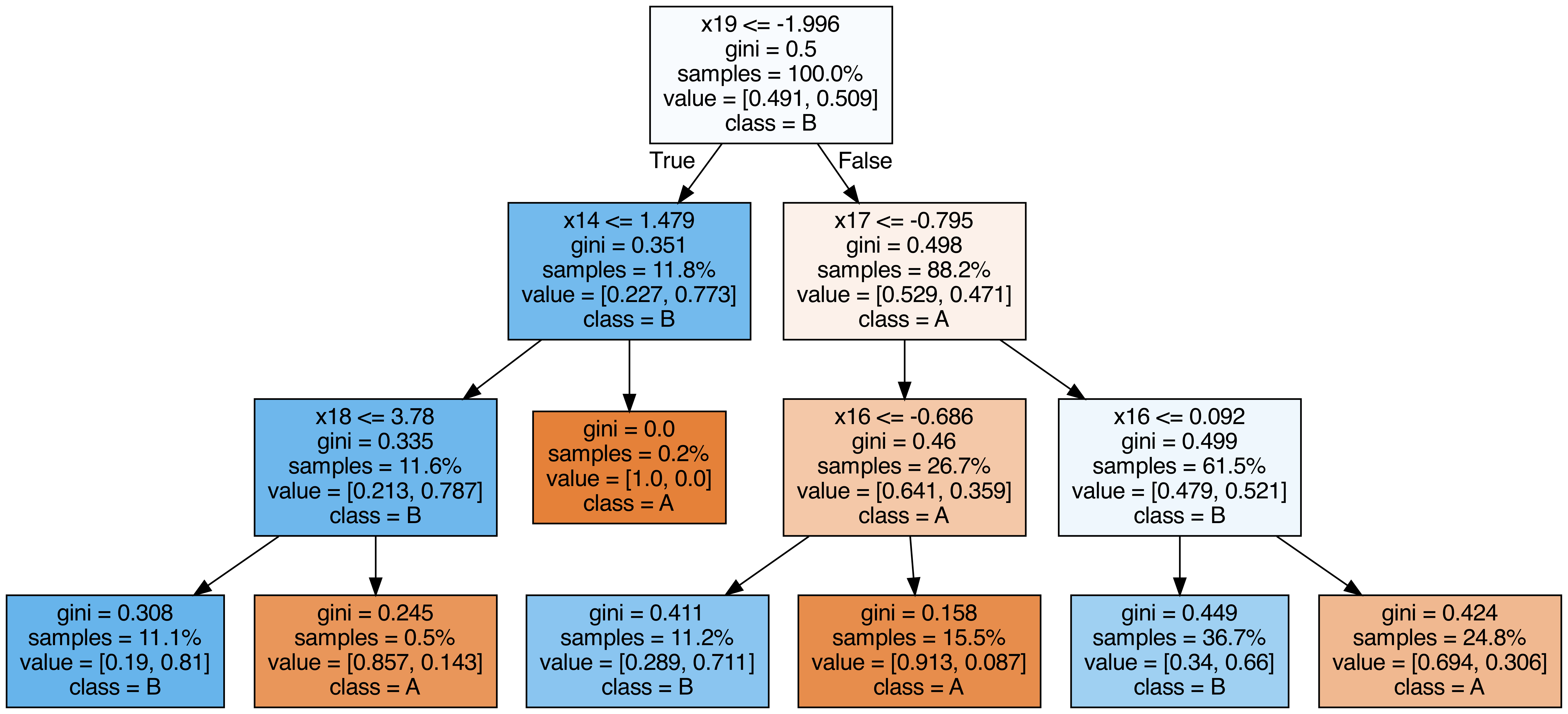
OOB (out-of-bag) Score #
We can confirm that the OOB and test data results are close to each other. Compare the OOB accuracies with the test data while changing the random numbers and tree depth.
from sklearn.metrics import accuracy_score
for i in range(10):
model_i = RandomForestClassifier(
n_estimators=50,
max_depth=3 + i % 2,
random_state=i,
bootstrap=True,
oob_score=True,
)
model_i.fit(X_train, y_train)
y_pred = model_i.predict(X_test)
oob_score = model_i.oob_score_
test_score = accuracy_score(y_test, y_pred)
print(f"OOB={oob_score} test={test_score}")
OOB=0.7868656716417910 test=0.8121212121212121
OOB=0.8101492537313433 test=0.8363636363636363
OOB=0.7886567164179105 test=0.8024242424242424
OOB=0.8161194029850747 test=0.8315151515151515
OOB=0.7910447761194029 test=0.8072727272727273
OOB=0.8101492537313433 test=0.833939393939394
OOB=0.7814925373134328 test=0.8133333333333334
OOB=0.8059701492537313 test=0.833939393939394
OOB=0.7832835820895523 test=0.7951515151515152
OOB=0.8083582089552239 test=0.8387878787878787