2.1.6
Bayesian Linear Regression
Summary
- Bayesian linear regression treats coefficients as random variables, estimating both predictions and their uncertainty.
- The posterior distribution is derived analytically from the prior and the likelihood, making the method robust for small or noisy datasets.
- The predictive distribution is Gaussian, so its mean and variance can be visualized and used for decision making.
BayesianRidgein scikit-learn automatically tunes the noise variance, which simplifies practical adoption.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
Mathematical formulation #
Assume a multivariate Gaussian prior with mean 0 and variance \(\tau^{-1}\) for the coefficient vector \(\boldsymbol\beta\), and Gaussian noise \(\epsilon_i \sim \mathcal{N}(0, \alpha^{-1})\) on the observations. The posterior becomes
$$ p(\boldsymbol\beta \mid \mathbf{X}, \mathbf{y}) = \mathcal{N}(\boldsymbol\beta \mid \boldsymbol\mu, \mathbf{\Sigma}) $$with
$$ \mathbf{\Sigma} = (\alpha \mathbf{X}^\top \mathbf{X} + \tau \mathbf{I})^{-1}, \qquad \boldsymbol\mu = \alpha \mathbf{\Sigma} \mathbf{X}^\top \mathbf{y}. $$The predictive distribution for a new input \(\mathbf{x}*\) is also Gaussian, \(\mathcal{N}(\hat{y}, \sigma_^2)\). BayesianRidge estimates \(\alpha\) and \(\tau\) from data, so you can use the model without hand-tuning them.
Experiments with Python #
The following example compares ordinary least squares with Bayesian linear regression on data containing outliers.
| |
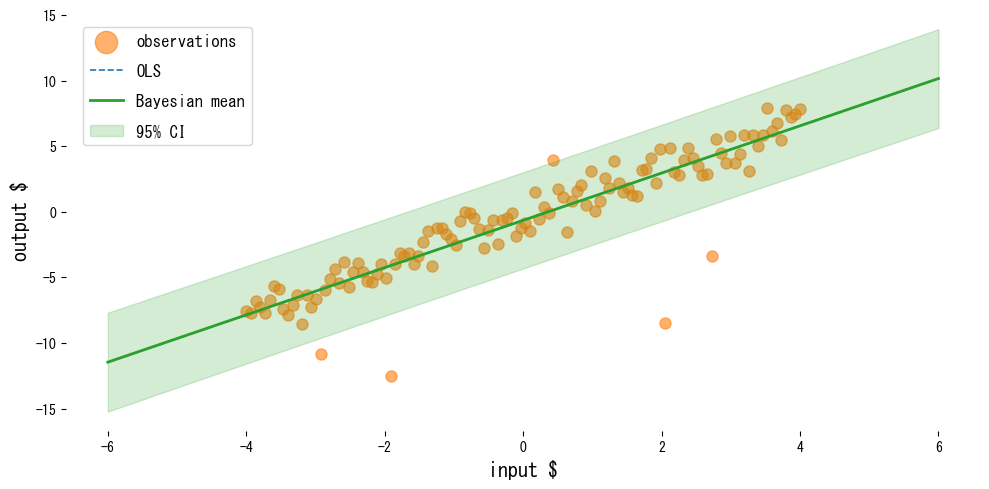
Reading the results #
- OLS is pulled toward the outliers, while Bayesian linear regression keeps the mean prediction more stable.
- Using
return_std=Trueyields the predictive standard deviation, which makes it easy to plot credible intervals. - Inspecting the posterior variance highlights which coefficients carry the most uncertainty.
References #
- Bishop, C. M. (2006). Pattern Recognition and Machine Learning. Springer.
- Murphy, K. P. (2012). Machine Learning: A Probabilistic Perspective. MIT Press.