2.1.1
Linear Regression
Summary
- Linear regression models the linear relationship between inputs and outputs and provides a baseline that is both predictive and interpretable.
- Ordinary least squares estimates the coefficients by minimizing the sum of squared residuals, yielding a closed-form solution.
- The slope tells us how much the output changes when the input increases by one unit, while the intercept represents the expected value when the input is zero.
- When noise or outliers are large, consider standardization and robust variants so that preprocessing and evaluation remain reliable.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
Mathematical formulation #
A univariate linear model is written as
$$ y = w x + b. $$By minimizing the sum of squared residuals \(\epsilon_i = y_i - (w x_i + b)\)
$$ L(w, b) = \sum_{i=1}^{n} \big(y_i - (w x_i + b)\big)^2, $$we obtain the analytic solution
$$ w = \frac{\sum_{i=1}^{n} (x_i - \bar{x})(y_i - \bar{y})}{\sum_{i=1}^{n} (x_i - \bar{x})^2}, \qquad b = \bar{y} - w \bar{x}, $$where \(\bar{x}\) and \(\bar{y}\) are the means of \(x\) and \(y\). The same idea extends to multivariate regression with vectors and matrices.
Experiments with Python #
The following code fits a simple regression line with scikit-learn and plots the result. The code is identical to the Japanese page so figures will match across languages.
| |
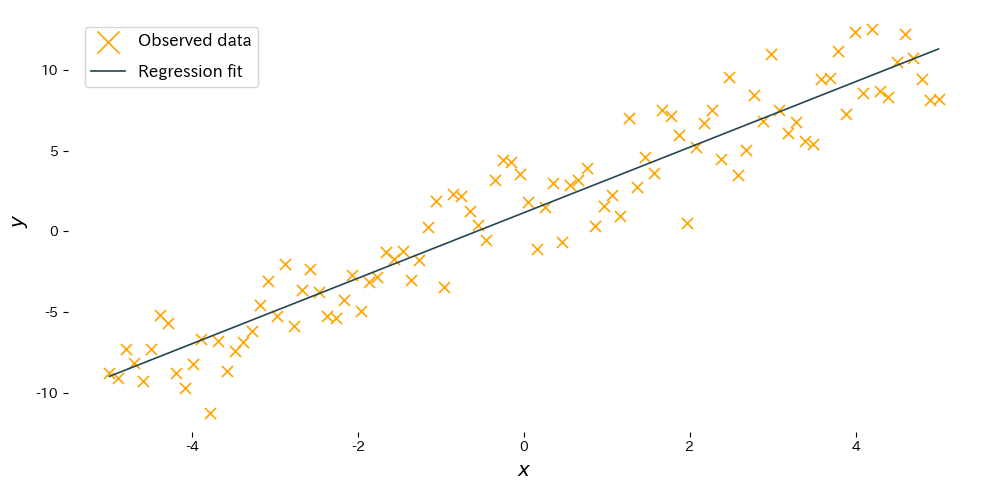
Reading the results #
- Slope \(w\): indicates how much the output increases or decreases when the input grows by one unit. The estimate should be close to the true slope.
- Intercept \(b\): shows the expected output when the input is 0, adjusting the vertical position of the line.
- Standardizing the features with
StandardScalerstabilizes learning when inputs vary in scale.
References #
- Draper, N. R., & Smith, H. (1998). Applied Regression Analysis (3rd ed.). John Wiley & Sons.
- Hastie, T., Tibshirani, R., & Friedman, J. (2009). The Elements of Statistical Learning. Springer.