2.1.9
Partial Least Squares Regression (PLS)
Summary
- Partial Least Squares (PLS) extracts latent factors that maximize the covariance between predictors and the target before performing regression.
- Unlike PCA, the learned axes incorporate target information, preserving predictive performance while reducing dimensionality.
- Tuning the number of latent factors stabilizes models in the presence of strong multicollinearity.
- Inspecting loadings reveals which combinations of features are most related to the target.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
Mathematical formulation #
Given a predictor matrix \(\mathbf{X}\) and response vector \(\mathbf{y}\), PLS alternates updates of latent scores \(\mathbf{t} = \mathbf{X} \mathbf{w}\) and \(\mathbf{u} = \mathbf{y} c\) so that their covariance \(\mathbf{t}^\top \mathbf{u}\) is maximized. Repeating this procedure yields a set of latent factors on which a linear regression model
$$ \hat{y} = \mathbf{t} \boldsymbol{b} + b_0 $$is fitted. The number of factors \(k\) is typically chosen via cross-validation.
Experiments with Python #
We compare PLS performance for different numbers of latent factors on the Linnerud fitness dataset.
| |
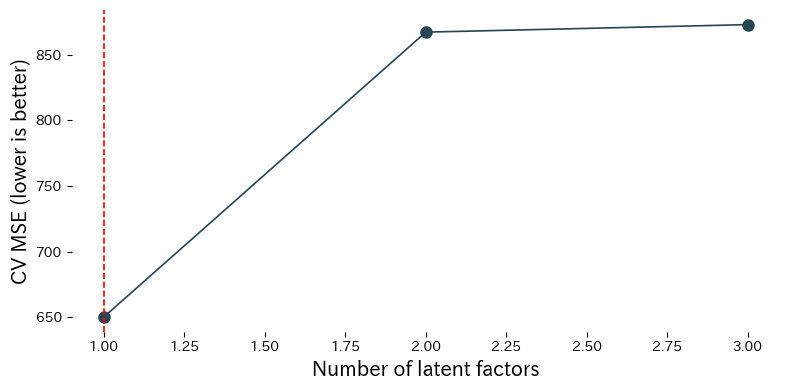
Reading the results #
- Cross-validated MSE decreases as factors are added, reaches a minimum, and then worsens if you keep adding more.
- Inspecting
x_loadings_andy_loadings_shows which features contribute most to each latent factor. - Standardizing inputs ensures features measured on different scales contribute evenly.
References #
- Wold, H. (1975). Soft Modelling by Latent Variables: The Non-Linear Iterative Partial Least Squares (NIPALS) Approach. In Perspectives in Probability and Statistics. Academic Press.
- Geladi, P., & Kowalski, B. R. (1986). Partial Least-Squares Regression: A Tutorial. Analytica Chimica Acta, 185, 1–17.