2.1.8
Principal Component Regression (PCR)
Summary
- Principal Component Regression (PCR) applies PCA to compress features before fitting linear regression, reducing instability from multicollinearity.
- Principal components prioritize directions with large variance, filtering noisy axes while preserving informative structure.
- Choosing how many components to keep balances overfitting risk and computational cost.
- Proper preprocessing—standardization and handling missing values—lays the groundwork for accuracy and interpretability.
Intuition #
This method should be interpreted through its assumptions, data conditions, and how parameter choices affect generalization.
Detailed Explanation #
Mathematical formulation #
Apply PCA to the standardized design matrix \(\mathbf{X}\) and retain the top \(k\) eigenvectors. With principal component scores \(\mathbf{Z} = \mathbf{X} \mathbf{W}_k\), the regression model
$$ y = \boldsymbol{\gamma}^\top \mathbf{Z} + b $$is learned. Coefficients in the original feature space are recovered via \(\boldsymbol{\beta} = \mathbf{W}_k \boldsymbol{\gamma}\). The number of components \(k\) is selected using cumulative explained variance or cross-validation.
Experiments with Python #
We evaluate cross-validation scores of PCR on the diabetes dataset as we vary the number of components.
| |
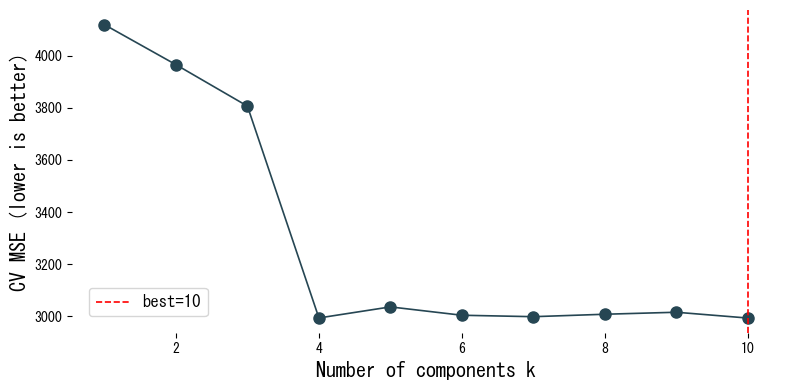
Reading the results #
- As the number of components increases, the training fit improves, but cross-validated MSE reaches a minimum at an intermediate value.
- Inspecting the explained variance ratio reveals how much of the overall variability each component captures.
- Component loadings indicate which original features contribute most to each principal direction.
References #
- Jolliffe, I. T. (2002). Principal Component Analysis (2nd ed.). Springer.
- Massy, W. F. (1965). Principal Components Regression in Exploratory Statistical Research. Journal of the American Statistical Association, 60(309), 234–256.