4.1.2
Stratified k-fold cross-validation
Summary
- Stratified k-fold preserves class proportions in every fold, which is essential for imbalanced datasets.
- Compare stratified and standard k-fold to visualise how class bias differs between them.
- Review design tips for extreme imbalance scenarios and how to interpret the results in practice.
- Cross-Validation — understanding this concept first will make learning smoother
| |
Building a model and running cross-validation #
Experimental dataset #
| |
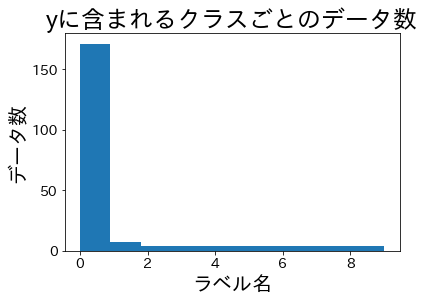
Class ratios after splitting #
Let’s split the data and verify the class proportions in the training and validation folds.
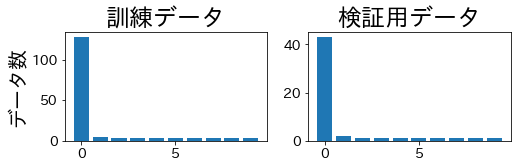
StratifiedKFold #
The class balance is maintained across both training and validation folds.
| |
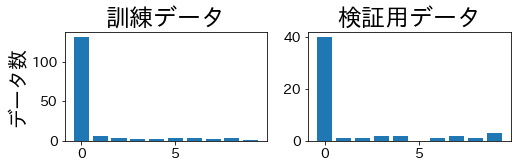
KFold #
Standard k-fold can produce validation folds that completely miss some minority classes.
| |
Practical considerations #
- Extreme imbalance: when minority classes have only a handful of samples, consider combining stratification with repeated cross-validation to reduce variance further.
- Regression tasks: use
StratifiedKFoldon discretised targets (binning) when cross-validation needs balanced target ranges. - Shuffle policy: set
shuffle=True(with a fixed random seed) when the dataset has temporal or grouped ordering that might bias folds.
Stratified k-fold is a drop-in replacement for standard k-fold when class balance matters. It produces fairer validation splits, stabilises metrics such as ROC-AUC, and improves comparability among models trained on imbalanced datasets.