4.2.1
Correlation coefficient
Summary- Understand the fundamentals of this metric, what it evaluates, and how to interpret the results.
- Compute and visualise the metric with Python 3.13 code examples, covering key steps and practical checkpoints.
- Combine charts and complementary metrics for effective model comparison and threshold tuning.
Correlation coefficient measures the strength of a linear relationship between two data or random variables.
It is an indicator that allows us to check whether there is a trend change of linear form between two variables, which can be expressed in the following equation.
$
\frac{\Sigma_{i=1}^N (x_i - \bar{x})(y_i - \bar{y})}{\sqrt{\Sigma_{i=1}^N(x_i - \bar{x})^2 \Sigma_{i=1}^N(y_i - \bar{y})^2 }}
$
It has the following properties
If correlation coefficient is close to 1, \(x\) increases → \(y\) also increases
The value of correlation coefficient does not change when \(x, y\) are multiplied by a low number
Calculate the correlation coefficient between two numerical columns
#
1
2
3
| import numpy as np
np.random.seed(777) # to fix random numbers
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| import matplotlib.pyplot as plt
import numpy as np
x = [xi + np.random.rand() for xi in np.linspace(0, 100, 40)]
y = [yi + np.random.rand() for yi in np.linspace(1, 50, 40)]
plt.figure(figsize=(5, 5))
plt.scatter(x, y)
plt.show()
coef = np.corrcoef(x, y)
print(coef)
|
[[1. 0.99979848]
[0.99979848 1. ]]
Collectively compute the correlation coefficient between multiple variables
#
1
2
3
4
5
| import seaborn as sns
df = sns.load_dataset("iris")
df.head()
|
<tr style="text-align: right;">
<th></th>
<th>sepal_length</th>
<th>sepal_width</th>
<th>petal_length</th>
<th>petal_width</th>
<th>species</th>
</tr>
<tr>
<th>0</th>
<td>5.1</td>
<td>3.5</td>
<td>1.4</td>
<td>0.2</td>
<td>setosa</td>
</tr>
<tr>
<th>1</th>
<td>4.9</td>
<td>3.0</td>
<td>1.4</td>
<td>0.2</td>
<td>setosa</td>
</tr>
<tr>
<th>2</th>
<td>4.7</td>
<td>3.2</td>
<td>1.3</td>
<td>0.2</td>
<td>setosa</td>
</tr>
<tr>
<th>3</th>
<td>4.6</td>
<td>3.1</td>
<td>1.5</td>
<td>0.2</td>
<td>setosa</td>
</tr>
<tr>
<th>4</th>
<td>5.0</td>
<td>3.6</td>
<td>1.4</td>
<td>0.2</td>
<td>setosa</td>
</tr>
Check the CORRELATION COEFFICIENCES between all variables
#
Using the iris dataset, let’s look at the correlation between variables.
1
| df.corr().style.background_gradient(cmap="YlOrRd")
|
<tr>
<th class="blank level0" > </th>
<th class="col_heading level0 col0" >sepal_length</th>
<th class="col_heading level0 col1" >sepal_width</th>
<th class="col_heading level0 col2" >petal_length</th>
<th class="col_heading level0 col3" >petal_width</th>
</tr>
<tr>
<th id="T_dbd76_level0_row0" class="row_heading level0 row0" >sepal_length</th>
<td id="T_dbd76_row0_col0" class="data row0 col0" >1.000000</td>
<td id="T_dbd76_row0_col1" class="data row0 col1" >-0.117570</td>
<td id="T_dbd76_row0_col2" class="data row0 col2" >0.871754</td>
<td id="T_dbd76_row0_col3" class="data row0 col3" >0.817941</td>
</tr>
<tr>
<th id="T_dbd76_level0_row1" class="row_heading level0 row1" >sepal_width</th>
<td id="T_dbd76_row1_col0" class="data row1 col0" >-0.117570</td>
<td id="T_dbd76_row1_col1" class="data row1 col1" >1.000000</td>
<td id="T_dbd76_row1_col2" class="data row1 col2" >-0.428440</td>
<td id="T_dbd76_row1_col3" class="data row1 col3" >-0.366126</td>
</tr>
<tr>
<th id="T_dbd76_level0_row2" class="row_heading level0 row2" >petal_length</th>
<td id="T_dbd76_row2_col0" class="data row2 col0" >0.871754</td>
<td id="T_dbd76_row2_col1" class="data row2 col1" >-0.428440</td>
<td id="T_dbd76_row2_col2" class="data row2 col2" >1.000000</td>
<td id="T_dbd76_row2_col3" class="data row2 col3" >0.962865</td>
</tr>
<tr>
<th id="T_dbd76_level0_row3" class="row_heading level0 row3" >petal_width</th>
<td id="T_dbd76_row3_col0" class="data row3 col0" >0.817941</td>
<td id="T_dbd76_row3_col1" class="data row3 col1" >-0.366126</td>
<td id="T_dbd76_row3_col2" class="data row3 col2" >0.962865</td>
<td id="T_dbd76_row3_col3" class="data row3 col3" >1.000000</td>
</tr>
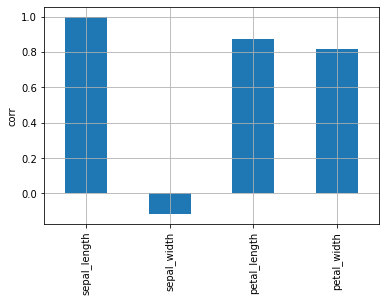
In the heatmap, it is hard to see where the correlation is highest. Check the bar chart to see which variables have the highest correlation with sepal_length.
1
| df.corr()["sepal_length"].plot.bar(grid=True, ylabel="corr")
|
When correlation coefficient is low
#
Check the data distribution when the correlation coefficient is low and confirm that the correlation coefficient may be low even when there is a relationship between variables.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
| n_samples = 1000
plt.figure(figsize=(12, 12))
for i, ci in enumerate(np.linspace(-1, 1, 16)):
ci = np.round(ci, 4)
mean = np.array([0, 0])
cov = np.array([[1, ci], [ci, 1]])
v1, v2 = np.random.multivariate_normal(mean, cov, size=n_samples).T
plt.subplot(4, 4, i + 1)
plt.plot(v1, v2, "x")
plt.title(f"r={ci}")
plt.tight_layout()
plt.show()
|
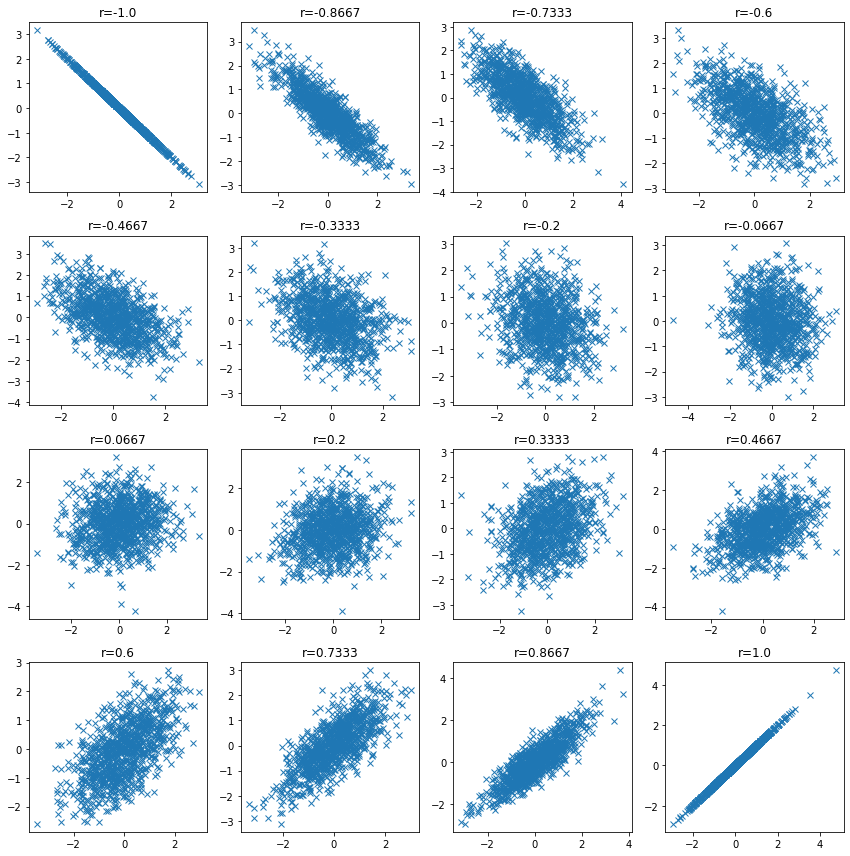
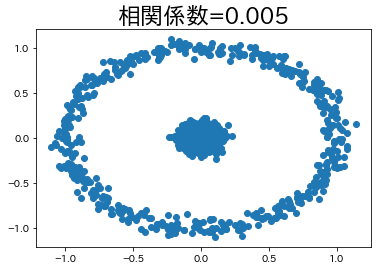
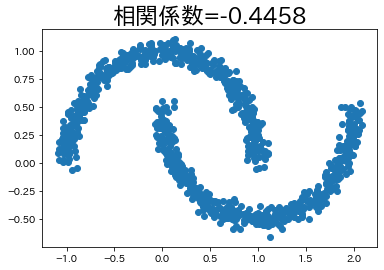
In some cases, there is a relationship between variables even if the correlation coefficient is low.
We will try to create such an example, albeit a simple one.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
| import japanize_matplotlib
from sklearn import datasets
japanize_matplotlib.japanize()
n_samples = 1000
circle, _ = datasets.make_circles(n_samples=n_samples, factor=0.1, noise=0.05)
moon, _ = datasets.make_moons(n_samples=n_samples, noise=0.05)
corr_circle = np.round(np.corrcoef(circle[:, 0], circle[:, 1])[1, 0], 4)
plt.title(f"correlation coefficient={corr_circle}", fontsize=23)
plt.scatter(circle[:, 0], circle[:, 1])
plt.show()
corr_moon = np.round(np.corrcoef(moon[:, 0], moon[:, 1])[1, 0], 4)
plt.title(f"correlation coefficient={corr_moon}", fontsize=23)
plt.scatter(moon[:, 0], moon[:, 1])
plt.show()
|