3.3.3
Yeo-Johnson transformation
<p>The <b>Yeo-Johnson</b> power transformation is a Box-Cox style transform that can stabilise variance and make skewed numerical features look more Gaussian <em>even when the data contain zeros or negative values</em>.</p>
Definition #
For an observation (y) and power parameter (\lambda), the Yeo-Johnson transform (T_\lambda(y)) is defined piecewise:
$$ T_\lambda(y)= \begin{cases} \dfrac{(y + 1)^\lambda - 1}{\lambda}, & y \ge 0,\ \lambda \ne 0,\\\\ \log(y + 1), & y \ge 0,\ \lambda = 0,\\\\ -\dfrac{(1 - y)^{2 - \lambda} - 1}{2 - \lambda}, & y < 0,\ \lambda \ne 2,\\\\ -\log(1 - y), & y < 0,\ \lambda = 2. \end{cases} $$- When (\lambda = 1) the transform leaves the data unchanged.
- Positive observations are treated like a Box-Cox transform on (y + 1).
- Negative observations are reflected around zero, allowing the method to cope with sign changes.
- The inverse transform is defined by solving the same cases for (y); SciPy provides it via
scipy.stats.yeojohnson_inverse.
The parameter (\lambda) is usually estimated by maximising the log-likelihood of the transformed data under a normal model. SciPy exposes the maximum-likelihood estimate through yeojohnson_normmax.
I. Yeo and R.A. Johnson, “A New Family of Power Transformations to Improve Normality or Symmetry”, Biometrika 87(4), 2000.
Worked example #
| |
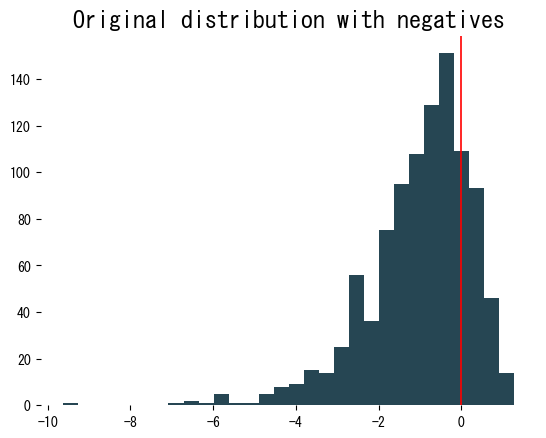
| |
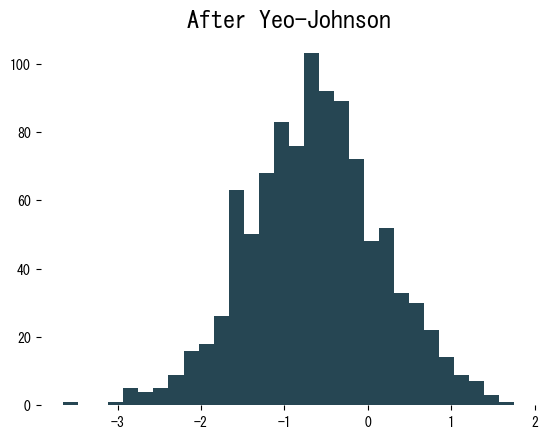
The histogram after transformation is much closer to symmetric. Because we explicitly pass lmbda, we can reuse the same parameter for validation or test data:
| |
Practical tips #
- Standardise features after the power transform if your model expects zero mean and unit variance.
- Apply the transformation parameters learned on the training split to every other split to avoid data leakage.
- When heavy tails remain, compare Yeo-Johnson with robust scalers such as
RobustScaler; combining them often works well.
This transformation is a drop-in replacement for Box-Cox in preprocessing pipelines that cannot assume strictly positive data.